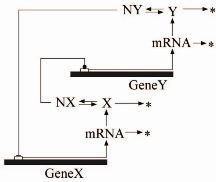
|
Systems biology seeks to understand living organisms by
modeling and analyzing the complex interactions of genes,
proteins, and other cell elements. These interactions occur
through biochemical reactions that take place inside the
cell or close to the cell membrane. Particularly crucial are
the chemical reactions that participate in the complex
regulatory mechanism that control cell functions such as the
heat shock response, which protects a cell against
environmental stresses (heat, cold, oxygen deprivation,
etc.); apoptosis, which leads to a programmed cell death
with minimal harm to nearby cells; chemotaxis, which permits
a cell to move in search of food or to flee from poisons; or
cell division, which results in two daughter cells from a
single parent cell.
Ultimately, the goal of systems biology is to transform
the methodology used for drug discovery, which is currently
dominated by mass experimentation. By enlarge, when faced
with a new disease or condition, drug developers
expose compromised cell cultures to a large number of
chemical compounds in the hope of finding a substance that
"treats" the disease. Finding such a substance, triggers a
second phase of experiments aimed at making sure that this
substance does not harm individual cells or organs. In
addition, a mechanism must be found to deliver the treatment
to the right cells. The goal of systems biology is to guide
this effort so that most effort is spent searching among the
most promising types of substances and making sure that all
cell functions that could be affected by the potential
treatment are not negatively affected.
What makes finding cures for diseases especially
challenging is the fact that cells are exquisitely regulated
mechanisms with multiple feedback loops. Suppose for example
that it is discovered that a particular disease develops
because a set of cells is lacking protein X. A naive cure
would be to inject X into the blood stream in an attempt to
increase its concentration. However, this can actually have
a completely opposite effect if the body interprets the high
concentration of X in the blood as a signal that this
protein is being overproduced and shuts down the natural
production of X. This is not unlike the apparent paradox
that results from placing a heater next to the temperature
feedback sensor of a central heating systems and suddenly
realizing that the whole building got much colder.
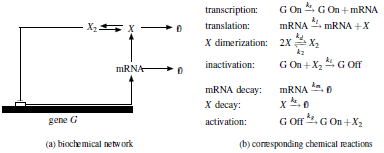 | Simple
gene regulatory network, where a gene G produces mRNA
(transcription), which in turn produces a protein X
(translation). The regulation of the production of X is
achieved by a negative feedback loop that results from
the fact that the protein X dimerizes to produce X2,
which acts as a repressor transcription factor that
inhibits transcription by preventing the RNA polymerase
from binding to the gene G, thus effectively
inactivating it by preventing the production of mRNA. To
the right-hand side of the network, we see a set of
chemical reactions that can embody this mechanism.
|
The goal of our research has been to develop tools to
analyze complex networks of biochemical reactions. Motivated
by the above observations, we are especially interested in
constructing dynamical models that highlight the feedback
mechanisms in cell regulation and that provide a qualitative
and quantitative understanding of how the different genes,
proteins, and other cell elements contribute to the observed
behavior (phenotype).
Gene regulatory mechanisms typically involve a large
number of distinct chemical species, but it is common for
some of these species to be represented by just a few
molecules, which can invalidate models based on the
deterministic chemical rate equation. Our work has been
using tools developed for Stochastic Hybrid Systems to
construct differential equations that accurately model the
stochastic effects present in biochemical networks.
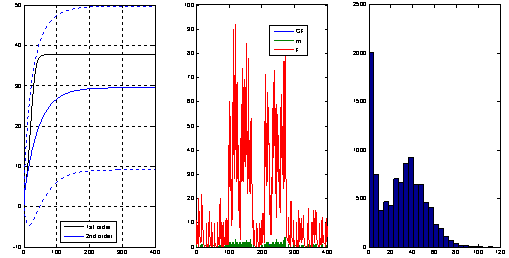 |
The left plot shows the evolution of the mean and
variance of the number of molecules of a particular
chemical species involved in a bio-chemical network
obtained from an ODE that approximates the moment
dynamics. In the middle plot we see a typical sample
path obtained through a Monte Carlo simulation and in
the right plot an histogram obtained from a large number
of such simulations. The moment dynamics ODE model was
obtained using the package StochDynTools
developed by our team and the Monte Carlo simulations
were obtained with Petzold's
StochKit.
|
Publications on this work can be found at the following
URL:
http://www.ece.ucsb.edu/~hespanha/published.html#13Biology
Software to compute moment dynamics can be found at the
following URL:
http://www.ece.ucsb.edu/~hespanha/software/stochdyntool.html
|